Role of PNFs in retroviral gene transfer
Simulations of how PNFs aid retroviral gene transfer
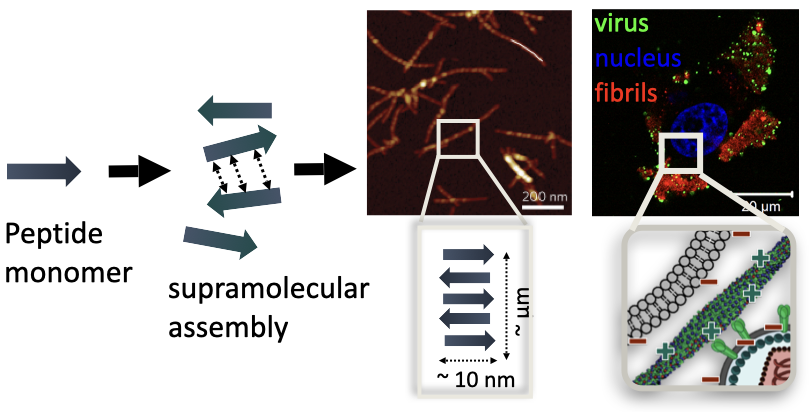
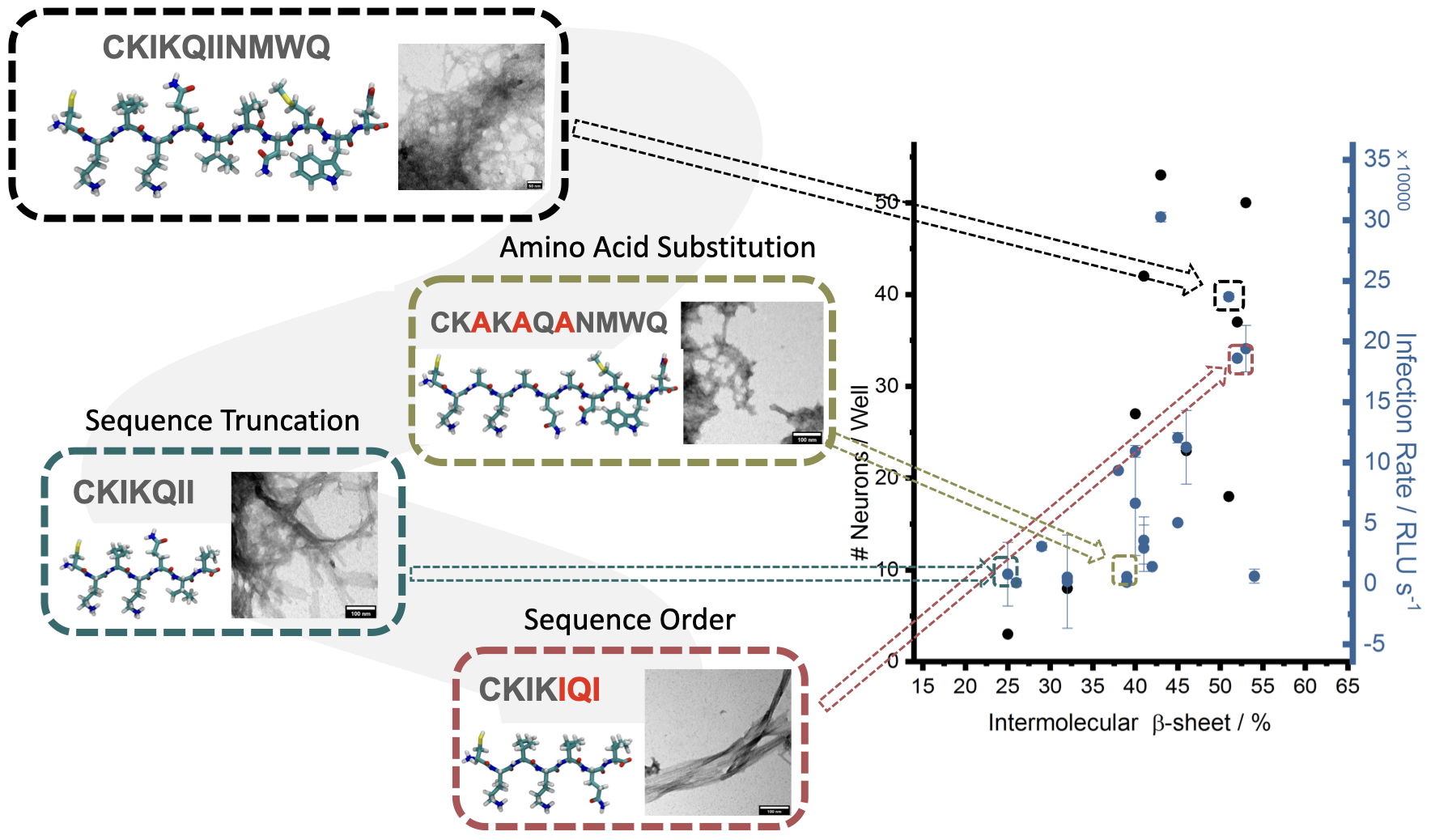
The chemical structure and morphology (self-assembly and amyloid formation) can affect the biological and materials properties of the peptide nanofibers. The reason why these nanofibers work so efficiently in retroviral gene transfer is attributed to the fact that positive charged amino acids (i.e. lysine) can interact with the negatively charged cell membranes and viral membranes, bringing them in close proximity and increasing the chances of gene transfer. However, there is no clear, microscopic evidence of how these fibers interact with the cell and viral membranes. Furthermore, the design of these materials rely on inefficient trial and error processes, where the peptide sequence (~ 10 amino acids long) can be any combination of the 20 possible naturally-occuring amino acids. When these short peptides fold in through intermolecular hydrogen bonding interactions between amino acids, different secondary structures, such as highly ordered β-sheets, a-helix or random coil, are formed. These molecular interactions between peptides have direct consequences on the morphology of the material, such as whether they form fibrils, micelles or aggregates, which then translate to materials properties, such as their stability and resistance, and biological activity in retroviral gene transfer applications. Past studies have found direct correlations between the secondary structure of nanofibrils and efficiency in retroviral gene transfer. Therefore, understanding of the relationship between a material’s microstructure at the atomic and molecular level and its properties will lead to rational, efficient and economical materials discoveries.
Finally, in order to understand the relationship between morphology and biological activity, we will perform simulation of the self-assembled fibers in solution in close proximity to cell membranes. With these simulations we hope to gain insight on which amino acids interact most with the cell membrane. Does the placement of these amino acids in the peptide sequence (primary structure), directly affect the final observed property (enhancement of retroviral gene transfer)? Do the fibers remain stable when interacting with the cell membranes? Are the fibrils degraded after transduction or stored as fibrils? By utilizing large scale MD simulations comparing cell protrusions/cell membranes interacting with the PNFs, we aim to answer such questions on PNF morphology during the transduction, binding sites on surface of PNFs, etc.